遗传算法与深度学习实战——利用进化计算优化深度学习模型
遗传算法与深度学习实战——利用进化计算优化深度学习模型
- 0. 前言
- 1. 利用进化计算优化深度学习模型
- 2. 利用进化策略优化深度学习模型
- 3. 利用差分计算优化深度学习模型
- 相关链接
0. 前言
我们已经学习了使用进化策略 (Evolutionary Strategies, ES) 和差分进化 (Differential Evolution, DE) 调整超参数,并取得了不错的结果。在本节中,我们将应用 ES 和 DE 作为神经进化优化器,并且继续使用圆圈或月亮数据集。
1. 利用进化计算优化深度学习模型
我们已经通过遗传算法优化简单深度学习 ( Deep learning, DL) 网络的权重/参数,除了遗传算法之外,我们还有更强大的进化方法,比如进化策略 (Evolutionary Strategies, ES) 和差分进化 (Differential Evolution, DE),它们可能表现更好。接下来,我们将研究这两种更高级的进化方法。
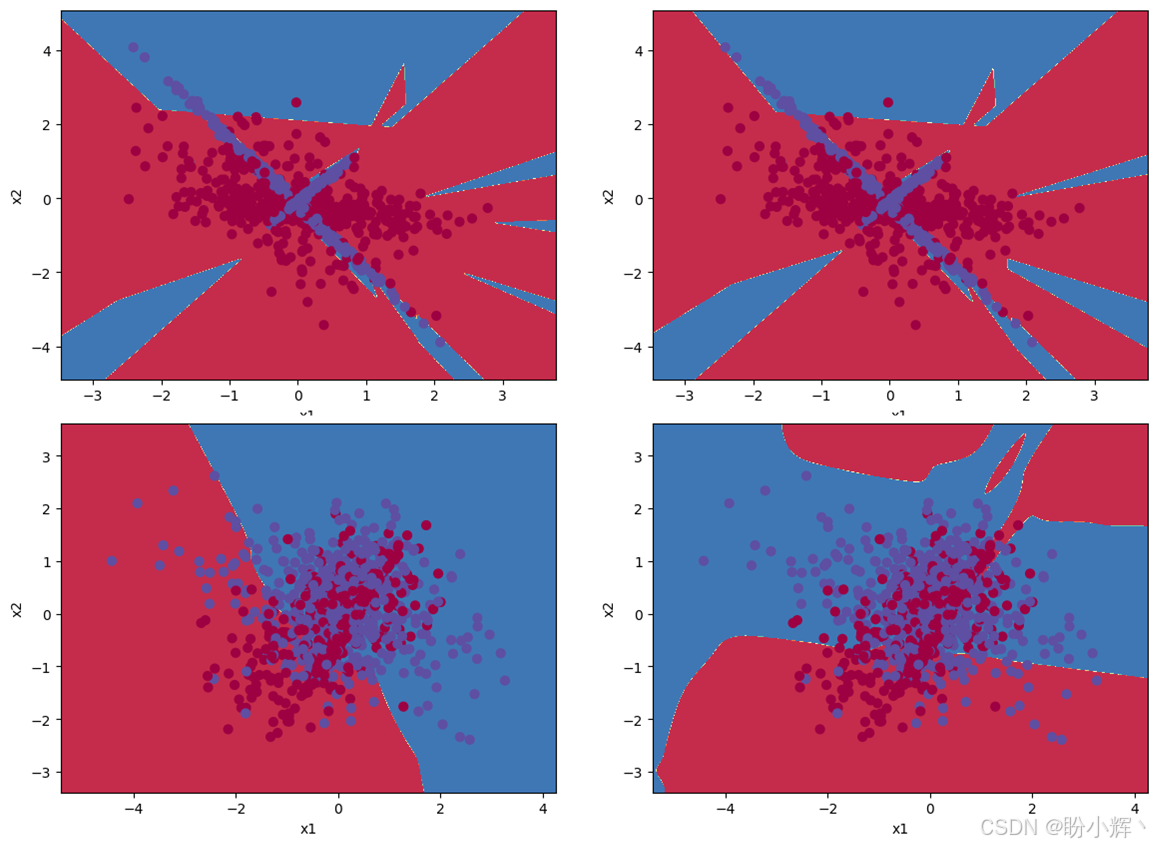
下图显示了使用 ES 和 DE 算法的运行结果,并观察 DE 和 ES 如何演化在每个数据集的权重。可以看到,ES不仅在解决困难数据集方面表现出色,而且还有解决更复杂问题的潜力。
2. 利用进化策略优化深度学习模型
在代码中,将遗传算法转换为进化策略 (Evolutionary Strategies, ES) 和差分进化 (Differential Evolution, DE) 算法。首先使用 ES 算法:
import numpy as np
import sklearn
import sklearn.datasets
import sklearn.linear_model
import matplotlib.pyplot as plt
from IPython.display import clear_outputfrom deap import algorithms
from deap import base
from deap import benchmarks
from deap import creator
from deap import toolsimport randomnumber_samples = 1000 #@param {type:"slider", min:100, max:1000, step:25}
difficulty = 5 #@param {type:"slider", min:1, max:5, step:1}
problem = "classification" #@param ["classification", "blobs", "gaussian quantiles", "moons", "circles"]
number_features = 2
number_classes = 2
middle_layer = 25 #@param {type:"slider", min:5, max:25, step:1}def load_data(problem): if problem == "classification":clusters = 1 if difficulty < 3 else 2informs = 1 if difficulty < 4 else 2data = sklearn.datasets.make_classification(n_samples = number_samples,n_features=number_features, n_redundant=0, class_sep=1/difficulty,n_informative=informs, n_clusters_per_class=clusters)if problem == "blobs":data = sklearn.datasets.make_blobs(n_samples = number_samples,n_features=number_features, centers=number_classes,cluster_std = difficulty)if problem == "gaussian quantiles":data = sklearn.datasets.make_gaussian_quantiles(mean=None, cov=difficulty,n_samples=number_samples,n_features=number_features,n_classes=number_classes,shuffle=True,random_state=None)if problem == "moons":data = sklearn.datasets.make_moons(n_samples = number_samples)if problem == "circles":data = sklearn.datasets.make_circles(n_samples = number_samples)return datadata = load_data(problem)
X, Y = data# Input Data
plt.figure("Input Data")
plt.scatter(X[:, 0], X[:, 1], c=Y, s=40, cmap=plt.cm.Spectral)def show_predictions(model, X, Y, name=""):""" display the labeled data X and a surface of prediction of model """x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.01), np.arange(y_min, y_max, 0.01))X_temp = np.c_[xx.flatten(), yy.flatten()]Z = model.predict(X_temp)plt.figure("Predictions " + name)plt.contourf(xx, yy, Z.reshape(xx.shape), cmap=plt.cm.Spectral)plt.ylabel('x2')plt.xlabel('x1')
plt.scatter(X[:, 0], X[:, 1],c=Y, s=40, cmap=plt.cm.Spectral)clf = sklearn.linear_model.LogisticRegressionCV()
clf.fit(X, Y)show_predictions(clf, X, Y, "Logistic regression")LR_predictions = clf.predict(X)
print("Logistic Regression accuracy : ", np.sum(LR_predictions == Y) / Y.shape[0])def sigmoid(x):return 1.0 / (1.0 + np.exp(-x)) ## Neural Network
class Neural_Network:def __init__(self, n_in, n_hidden, n_out):# Network dimensionsself.n_x = n_inself.n_h = n_hiddenself.n_y = n_out# Parameters initializationself.W1 = np.random.randn(self.n_h, self.n_x) * 0.01self.b1 = np.zeros((self.n_h, 1))self.W2 = np.random.randn(self.n_y, self.n_h) * 0.01self.b2 = np.zeros((self.n_y, 1))self.parameters = [self.W1, self.b1, self.W2, self.b2]def forward(self, X):""" Forward computation """self.Z1 = self.W1.dot(X.T) + self.b1self.A1 = np.tanh(self.Z1)self.Z2 = self.W2.dot(self.A1) + self.b2self.A2 = sigmoid(self.Z2)def set_parameters(self, individual):"""Sets model parameters """idx = 0for p in self.parameters: size = p.sizesh = p.shapet = individual[idx:idx+size]t = np.array(t)t = np.reshape(t, sh)p -= pp += tidx += sizedef predict(self, X):""" Compute predictions with just a forward pass """self.forward(X)return np.round(self.A2).astype(np.int)nn = Neural_Network(2, middle_layer, 1)
number_of_genes = sum([p.size for p in nn.parameters])
print(number_of_genes)individual = np.ones(number_of_genes)
nn.set_parameters(individual)
print(nn.parameters)IND_SIZE = number_of_genes
MIN_VALUE = -1000
MAX_VALUE = 1000
MIN_STRATEGY = 0.5
MAX_STRATEGY = 5CXPB = .6
MUTPB = .3creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Individual", list, typecode="d", fitness=creator.FitnessMin, strategy=None)
creator.create("Strategy", list, typecode="d")def generateES(icls, scls, size, imin, imax, smin, smax): ind = icls(random.uniform(imin, imax) for _ in range(size)) ind.strategy = scls(random.uniform(smin, smax) for _ in range(size)) return inddef checkStrategy(minstrategy):def decorator(func):def wrappper(*args, **kargs):children = func(*args, **kargs)for child in children:for i, s in enumerate(child.strategy):if s < minstrategy:child.strategy[i] = minstrategyreturn childrenreturn wrappper
return decoratordef custom_blend(ind1, ind2, alpha): for i, (x1, s1, x2, s2) in enumerate(zip(ind1, ind1.strategy,ind2, ind2.strategy)):# Blend the valuesgamma = (1. + 2. * alpha) * random.random() - alphaind1[i] = (1. - gamma) * x1 + gamma * x2ind2[i] = gamma * x1 + (1. - gamma) * x2# Blend the strategiesgamma = (1. + 2. * alpha) * random.random() - alphaind1.strategy[i] = (1. - gamma) * s1 + gamma * s2ind2.strategy[i] = gamma * s1 + (1. - gamma) * s2
return ind1, ind2toolbox = base.Toolbox()
toolbox.register("individual", generateES, creator.Individual, creator.Strategy,IND_SIZE, MIN_VALUE, MAX_VALUE, MIN_STRATEGY, MAX_STRATEGY)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("mate", custom_blend, alpha=0.5)
toolbox.register("mutate", tools.mutESLogNormal, c=1.0, indpb=0.06)
toolbox.register("select", tools.selTournament, tournsize=5)toolbox.decorate("mate", checkStrategy(MIN_STRATEGY))
toolbox.decorate("mutate", checkStrategy(MIN_STRATEGY))nn = Neural_Network(2, middle_layer, 1)
nn.set_parameters(individual)
print(nn.parameters)show_predictions(nn, X, Y, "Neural Network")nn_predictions = nn.predict(X)
print("Neural Network accuracy : ", np.sum(nn_predictions == Y) / Y.shape[0])def evaluate(individual): nn.set_parameters(individual)nn_predictions = nn.predict(X)return 1/np.sum(nn_predictions == Y) / Y.shape[0], toolbox.register("evaluate", evaluate)MU = 340 #@param {type:"slider", min:5, max:1000, step:5}
LAMBDA = 1000 #@param {type:"slider", min:5, max:1000, step:5}
NGEN = 100 #@param {type:"slider", min:100, max:1000, step:10}
RGEN = 10 #@param {type:"slider", min:1, max:100, step:1}
CXPB = .6
MUTPB = .3random.seed(64)pop = toolbox.population(n=MU)
hof = tools.HallOfFame(1)
stats = tools.Statistics(lambda ind: ind.fitness.values)
stats.register("avg", np.mean)
stats.register("std", np.std)
stats.register("min", np.min)
stats.register("max", np.max)best = None
history = []for g in range(NGEN):pop, logbook = algorithms.eaMuCommaLambda(pop, toolbox, mu=MU, lambda_=LAMBDA, cxpb=CXPB, mutpb=MUTPB, ngen=RGEN, stats=stats, halloffame=hof, verbose=False)best = hof[0] clear_output()print(f"Gen ({(g+1)*RGEN})")show_predictions(nn, X, Y, "Neural Network") nn_predictions = nn.predict(X)print("Current Neural Network accuracy : ", np.sum(nn_predictions == Y) / Y.shape[0])plt.show()nn.set_parameters(best)show_predictions(nn, X, Y, "Best Neural Network")plt.show()nn_predictions = nn.predict(X)fitness = np.sum(nn_predictions == Y) / Y.shape[0]print("Best Neural Network accuracy : ", fitness)if fitness > .99: #stop conditionbreak
3. 利用差分计算优化深度学习模型
接下来,应用 DE 作为神经进化优化器:
import numpy as np
import sklearn
import sklearn.datasets
import sklearn.linear_model
import matplotlib.pyplot as plt
from IPython.display import clear_outputfrom deap import algorithms
from deap import base
from deap import benchmarks
from deap import creator
from deap import toolsimport random
import array
import timenumber_samples = 1000 #@param {type:"slider", min:100, max:1000, step:25}
difficulty = 5 #@param {type:"slider", min:1, max:5, step:1}
problem = "classification" #@param ["classification", "blobs", "gaussian quantiles", "moons", "circles"]
number_features = 2
number_classes = 2
middle_layer = 25 #@param {type:"slider", min:5, max:25, step:1}def load_data(problem): if problem == "classification":clusters = 1 if difficulty < 3 else 2informs = 1 if difficulty < 4 else 2data = sklearn.datasets.make_classification(n_samples = number_samples,n_features=number_features, n_redundant=0, class_sep=1/difficulty,n_informative=informs, n_clusters_per_class=clusters)if problem == "blobs":data = sklearn.datasets.make_blobs(n_samples = number_samples,n_features=number_features, centers=number_classes,cluster_std = difficulty)if problem == "gaussian quantiles":data = sklearn.datasets.make_gaussian_quantiles(mean=None, cov=difficulty,n_samples=number_samples,n_features=number_features,n_classes=number_classes,shuffle=True,random_state=None)if problem == "moons":data = sklearn.datasets.make_moons(n_samples = number_samples)if problem == "circles":data = sklearn.datasets.make_circles(n_samples = number_samples)return datadata = load_data(problem)
X, Y = data# Input Data
plt.figure("Input Data")
plt.scatter(X[:, 0], X[:, 1], c=Y, s=40, cmap=plt.cm.Spectral)def show_predictions(model, X, Y, name=""):""" display the labeled data X and a surface of prediction of model """x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.01), np.arange(y_min, y_max, 0.01))X_temp = np.c_[xx.flatten(), yy.flatten()]Z = model.predict(X_temp)plt.figure("Predictions " + name)plt.contourf(xx, yy, Z.reshape(xx.shape), cmap=plt.cm.Spectral)plt.ylabel('x2')plt.xlabel('x1')
plt.scatter(X[:, 0], X[:, 1],c=Y, s=40, cmap=plt.cm.Spectral)clf = sklearn.linear_model.LogisticRegressionCV()
clf.fit(X, Y)show_predictions(clf, X, Y, "Logistic regression")LR_predictions = clf.predict(X)
print("Logistic Regression accuracy : ", np.sum(LR_predictions == Y) / Y.shape[0])def sigmoid(x):return 1.0 / (1.0 + np.exp(-x)) ## Neural Network
class Neural_Network:def __init__(self, n_in, n_hidden, n_out):# Network dimensionsself.n_x = n_inself.n_h = n_hiddenself.n_y = n_out# Parameters initializationself.W1 = np.random.randn(self.n_h, self.n_x) * 0.01self.b1 = np.zeros((self.n_h, 1))self.W2 = np.random.randn(self.n_y, self.n_h) * 0.01self.b2 = np.zeros((self.n_y, 1))self.parameters = [self.W1, self.b1, self.W2, self.b2]def forward(self, X):""" Forward computation """self.Z1 = self.W1.dot(X.T) + self.b1self.A1 = np.tanh(self.Z1)self.Z2 = self.W2.dot(self.A1) + self.b2self.A2 = sigmoid(self.Z2)def set_parameters(self, individual):"""Sets model parameters """idx = 0for p in self.parameters: size = p.sizesh = p.shapet = individual[idx:idx+size]t = np.array(t)t = np.reshape(t, sh)p -= pp += tidx += sizedef predict(self, X):""" Compute predictions with just a forward pass """self.forward(X)return np.round(self.A2).astype(np.int)nn = Neural_Network(2, middle_layer, 1)
number_of_genes = sum([p.size for p in nn.parameters])
print(number_of_genes)individual = np.ones(number_of_genes)
nn.set_parameters(individual)
print(nn.parameters)NDIM = number_of_genes
CR = 0.25
F_ = 1creator.create("FitnessMin", base.Fitness, weights=(-1.0,))
creator.create("Individual", array.array, typecode='d', fitness=creator.FitnessMin)toolbox = base.Toolbox()
toolbox.register("attr_float", random.uniform, -1, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual, toolbox.attr_float, NDIM)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register("select", tools.selRandom, k=3)nn = Neural_Network(2, middle_layer, 1)
nn.set_parameters(individual)
print(nn.parameters)show_predictions(nn, X, Y, "Neural Network")nn_predictions = nn.predict(X)
print("Neural Network accuracy : ", np.sum(nn_predictions == Y) / Y.shape[0])def evaluate(individual): nn.set_parameters(individual)nn_predictions = nn.predict(X)return 1/np.sum(nn_predictions == Y) / Y.shape[0], toolbox.register("evaluate", evaluate)MU = 340 #@param {type:"slider", min:5, max:1000, step:5}
NGEN = 1000 #@param {type:"slider", min:100, max:1000, step:10}
RGEN = 10 #@param {type:"slider", min:1, max:10, step:1}random.seed(64)pop = toolbox.population(n=MU)
hof = tools.HallOfFame(1)
stats = tools.Statistics(lambda ind: ind.fitness.values)
stats.register("avg", np.mean)
stats.register("std", np.std)
stats.register("min", np.min)
stats.register("max", np.max)logbook = tools.Logbook()
logbook.header = "gen", "evals", "std", "min", "avg", "max"best = None
history = []# Evaluate the individuals
fitnesses = toolbox.map(toolbox.evaluate, pop)
for ind, fit in zip(pop, fitnesses):ind.fitness.values = fitrecord = stats.compile(pop)
logbook.record(gen=0, evals=len(pop), **record)
print(logbook.stream)
for g in range(1, NGEN):for k, agent in enumerate(pop):a,b,c = toolbox.select(pop)y = toolbox.clone(agent)index = random.randrange(NDIM)for i, value in enumerate(agent):if i == index or random.random() < CR:y[i] = a[i] + F_*(b[i]-c[i])y.fitness.values = toolbox.evaluate(y)if y.fitness > agent.fitness:pop[k] = yhof.update(pop) record = stats.compile(pop) best = hof[0]if ((g+1) % RGEN) == 0:clear_output()print(f"Gen ({(g+1)})")show_predictions(nn, X, Y, "Neural Network") nn_predictions = nn.predict(X)print("Current Neural Network accuracy : ", np.sum(nn_predictions == Y) / Y.shape[0])plt.show()nn.set_parameters(best)show_predictions(nn, X, Y, "Best Neural Network")plt.show()nn_predictions = nn.predict(X)fitness = np.sum(nn_predictions == Y) / Y.shape[0]print("Best Neural Network accuracy : ", fitness)if fitness > .99: #stop conditionbreaktime.sleep(1)
对于本节所用的样本数据集,简单的遗传算法方法通常表现最佳,DE 明显是不太理想的选择,而 ES 具有一定的潜力。可以通过完成以下问题进一步了解不同神经优化算法的应用场景:
- 调整超参数,观察它们对
DE或ES的影响。 - 调整特定的进化方法超参数——
ES的最小和最大策略,或DE的pmin/pmax和smin/smax
相关链接
遗传算法与深度学习实战(1)——进化深度学习
遗传算法与深度学习实战(4)——遗传算法(Genetic Algorithm)详解与实现
遗传算法与深度学习实战(14)——进化策略详解与实现
遗传算法与深度学习实战(15)——差分进化详解与实现
遗传算法与深度学习实战(16)——神经网络超参数优化
遗传算法与深度学习实战(19)——使用粒子群优化自动超参数优化
遗传算法与深度学习实战(20)——使用进化策略自动超参数优化
遗传算法与深度学习实战(21)——使用差分搜索自动超参数优化
遗传算法与深度学习实战(22)——使用Numpy构建神经网络
遗传算法与深度学习实战(23)——利用遗传算法优化深度学习模型
