单细胞cluster/细胞亚群的标志识别工具—FindAllmarkers/presto/COSG/starTracer算法学习
随着细胞数量的增加,使用 FindAllMarkers进行标志基因分析的速度变得很慢,因此笔者就把几种常用的标志基因检测工具都用来试了试。
步骤流程
1.导入
rm(list = ls())
V5_path = "/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/seurat5/"
.libPaths(V5_path)
.libPaths()
library(Seurat) #v5
library(presto)
load("scRNA.Rdata")
dim(scRNA)
# [1] 24357 4031# check
DimPlot(scRNA,group.by = "celltype")
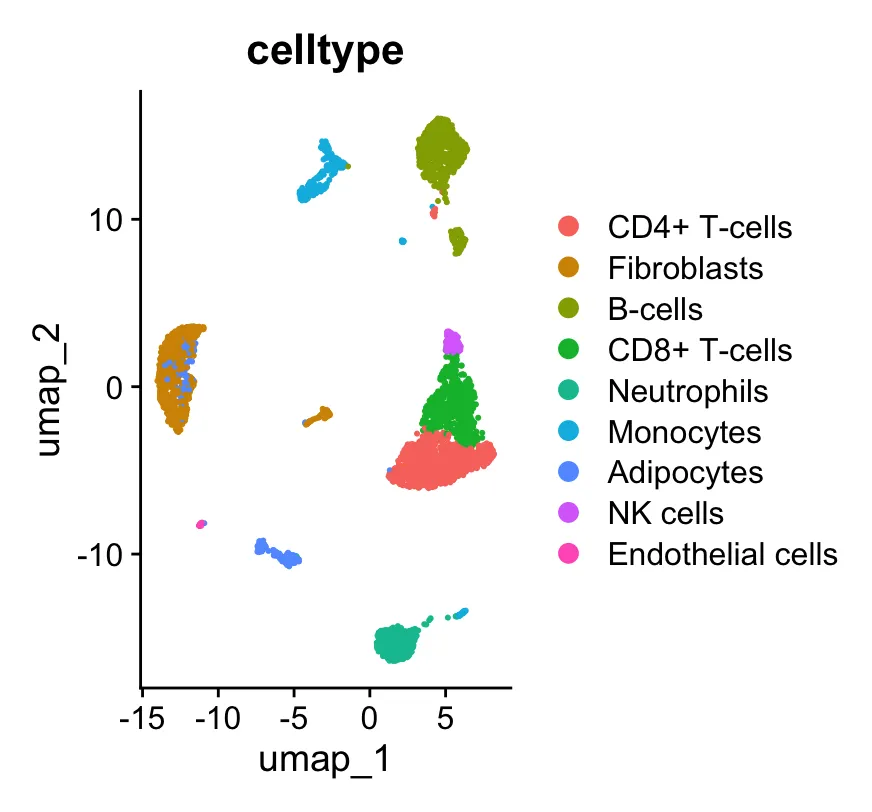
2.FindAllmarkers
start_time <- Sys.time() # 记录初始时间
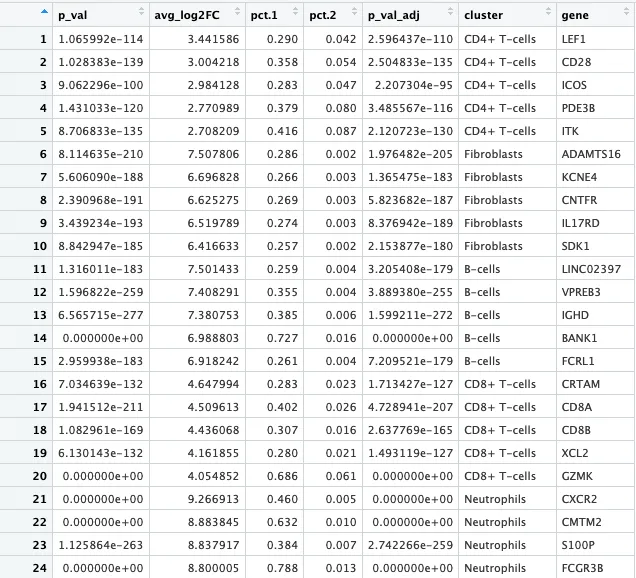
res <- FindAllMarkers(object = scRNA, only.pos = TRUE,min.pct = 0.25,logfc.threshold = 0.25)
end_time <- Sys.time() # 记录终止时间
end_time - start_time # 计算时间差
# Time difference of 3.996232 secs # top5
library(dplyr)
top5 <- res %>% group_by(cluster) %>% # 按照cluster分析arrange(desc(avg_log2FC), .by_group=T) %>%top_n(5, avg_log2FC) # 对每个分组选择avg_log2FC排名前5的基因
top5
运行时间是3.996232秒
logfc.threshold: log2倍数变化(log fold change, LogFC)的阈值。默认是0.1,
min.pct: 基因在至少一个群体中的表达比例阈值。默认是0.01,即基因需要在至少1%的细胞中表达。
only.pos: 是否只返回在给定群体中上调的基因,默认为 FALSE。
3.presto
library(presto)
start_time <- Sys.time() # 记录初始时间
# groups_use = c("B-cells", "T-cells") 如果附加了这个参数就是两群细胞之间比较
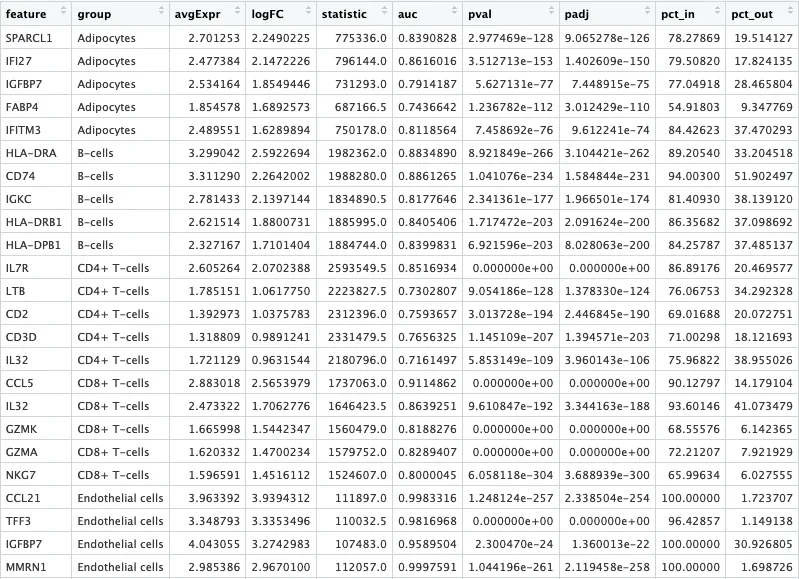
presto <- wilcoxauc(scRNA,group_by = "celltype")
end_time <- Sys.time() # 记录终止时间
end_time - start_time # 计算时间差
# Time difference of 0.8209939 secs presto_top5 <- presto %>%filter(logFC > 0.25 & pct_in > 25 & padj < 0.05) %>%group_by(group) %>%arrange(desc(logFC), .by_group=T) %>%top_n(n = 5, wt = logFC)
presto_top5
运行时间是0.8209939秒
看了这个函数的参数介绍,没有明确写出默认值,那就按照findallmarker的来。
4.COSG
library(COSG)
start_time <- Sys.time() # 记录初始时间
cosg <- cosg(scRNA,groups='all', #考虑全部分组assay='RNA',slot='data',mu=1, #惩罚项参数,值越大remove_lowly_expressed=TRUE, #是否过滤低表达基因expressed_pct=0.1, #设置阈值(规定百分比)n_genes_user=100 #每个cluster定义Top-N个marker gene
)
end_time <- Sys.time() # 记录终止时间
end_time - start_time # 计算时间差
# Time difference of 0.7045739 secs# Check the marker genes:
head(cosg$names)
# CD4+ T-cells Fibroblasts B-cells CD8+ T-cells Neutrophils Monocytes Adipocytes NK cells Endothelial cells
# 1 IL7R COL6A2 CD79A GZMK FCGR3B MS4A6A AQP1 FGFBP2 TFF3
# 2 CD28 C1S BANK1 CCL5 S100A8 CD163 PLVAP KLRF1 TBX1
# 3 MAL COL6A1 MS4A1 CD8A G0S2 MS4A7 ADGRL4 GNLY MMRN1
# 4 LEF1 C1R IGHM GZMA FPR1 CD68 PALMD KLRD1 CCL21
# 5 PDE3B SERPINF1 TNFRSF13C CD8B CSF3R CSF1R MMRN2 GZMB PROX1
# 6 TCF7 FBLN1 RALGPS2 CRTAM S100A9 C1QA VWF PRF1 NTS
head(cosg$scores)
# CD4+ T-cells Fibroblasts B-cells CD8+ T-cells Neutrophils Monocytes Adipocytes NK cells Endothelial cells
# 1 0.5226850 0.8841391 0.8285084 0.6263870 0.8532547 0.6944153 0.6827161 0.7548425 0.8540223
# 2 0.4225969 0.8769785 0.8008367 0.5764124 0.8392333 0.6699419 0.6639528 0.7526524 0.8356658
# 3 0.4225396 0.8598057 0.7772619 0.5161820 0.8104408 0.6528903 0.6603293 0.7123577 0.7992476
# 4 0.4191782 0.8500572 0.6866467 0.4842701 0.8070109 0.6226761 0.6008893 0.6192011 0.7812084
# 5 0.4044170 0.8455154 0.6860877 0.4538657 0.8032351 0.6146647 0.5992065 0.6186626 0.7576402
# 6 0.3970676 0.8436453 0.6035435 0.4133991 0.7707526 0.6091362 0.5956636 0.5936507 0.7520553
运行时间是0.7045739秒
COSG score取值范围为0~1。值越大,表示为cluster specific marker gene
5.starTracer
library(starTracer)
start_time <- Sys.time() # 记录初始时间
starTracer <- searchMarker(x = scRNA, # 输入一个Seurat对象、一个dgCMatrix或者是一个Average Expression Matrixthresh.1 = 0.5, # 建议使用默认值thresh.2 = 0.3, # thresh2越高,表达水平越高,特异性越低。请设置在0-1范围内method = "pos", # pos/all/negnum = 5, # 输出的top N marker基因数量gene.use = NULL, #留空使用所有基因,设置为“HVG”则使用HVGmeta.data = NULL,ident.use = NULL
)
end_time <- Sys.time() # 记录终止时间
end_time - start_time # 计算时间差
# Time difference of 1.751583 secs#您可以使用以下代码来获取marker基因的pseudo-bulk表达矩阵
starTracer$expr.use[starTracer$genes.markers,]#您可以使用以下代码来获取marker基因记忆其对应的cluster
starTracer$para_frame[starTracer$genes.markers,c("max.X","gene")]
# max.X gene
# SH2D1B NK cells SH2D1B
# KLRC2 NK cells KLRC2
# FGFBP2 NK cells FGFBP2
# KLRF1 NK cells KLRF1
# KIR2DL1 NK cells KIR2DL1
# TFF3 Endothelial cells TFF3
# GPR182 Endothelial cells GPR182
# STAB2 Endothelial cells STAB2
# NTS Endothelial cells NTS
# AC007998.3 Endothelial cells AC007998.3
# LYPD2 Monocytes LYPD2
# SIGLEC8 Monocytes SIGLEC8
# ASGR2 Monocytes ASGR2
# AL662860.1 Monocytes AL662860.1
# KCNA7 Monocytes KCNA7
# DLK1 Fibroblasts DLK1
# ADCY5 Fibroblasts ADCY5
# ADGRL3 Fibroblasts ADGRL3
# FAM43B Fibroblasts FAM43B
# CCDC8 Fibroblasts CCDC8
# S100P Neutrophils S100P
# CMTM2 Neutrophils CMTM2
# LINC00664 Neutrophils LINC00664
# PI3 Neutrophils PI3
# MGAM Neutrophils MGAM
# ELAVL2 Adipocytes ELAVL2
# GC Adipocytes GC
# AMBP Adipocytes AMBP
# TFF1 Adipocytes TFF1
# TFF2 Adipocytes TFF2
# IL17A CD4+ T-cells IL17A
# CCR8 CD4+ T-cells CCR8
# KLHL7-DT CD4+ T-cells KLHL7-DT
# AC026471.3 CD4+ T-cells AC026471.3
# IGFL2 CD4+ T-cells IGFL2
# LINC00461 CD8+ T-cells LINC00461
# AC026954.2 CD8+ T-cells AC026954.2
# AC023051.1 CD8+ T-cells AC023051.1
# TRDV2 CD8+ T-cells TRDV2
# RSPH6A CD8+ T-cells RSPH6A
# IGKV1-16 B-cells IGKV1-16
# LCN8 B-cells LCN8
# IGHV4-61 B-cells IGHV4-61
# IGHV1-24 B-cells IGHV1-24
# PLEKHG7 B-cells PLEKHG7
运行时间是1.751583秒
笔者觉得结果还是存在差异的hhh,使用哪一种大家仁者见仁智者见智吧~
参考资料:
-
Seurat:https://satijalab.org/seurat/articles/seurat5_integration
-
presto:https://github.com/immunogenomics/presto https://immunogenomics.github.io/presto/articles/getting-started.html
-
COSG:https://github.com/genecell/COSGR
-
starTracer:https://github.com/JerryZhang-1222/starTracer
-
单细胞天地:https://mp.weixin.qq.com/s/Y-NUJmkqcUWe01YVmIrjDA https://mp.weixin.qq.com/s/x0zk66873PQ3O1MYNXc0oA
-
生信技能树:https://mp.weixin.qq.com/s/vtpdxDDd-ZoW0mqfKiPsdg https://mp.weixin.qq.com/s/7GkGUIq5hB-fS27AyGjnrA
致谢:感谢曾老师以及生信技能树团队全体成员。
注:若对内容有疑惑或者有发现明确错误的朋友,请联系后台(欢迎交流)。更多内容可关注公众号:生信方舟
- END -
